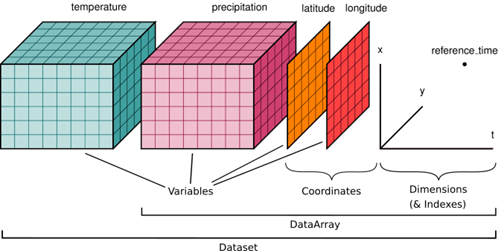
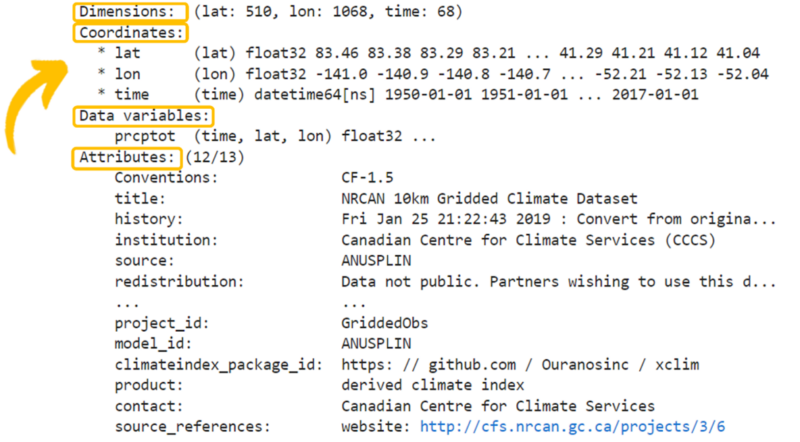
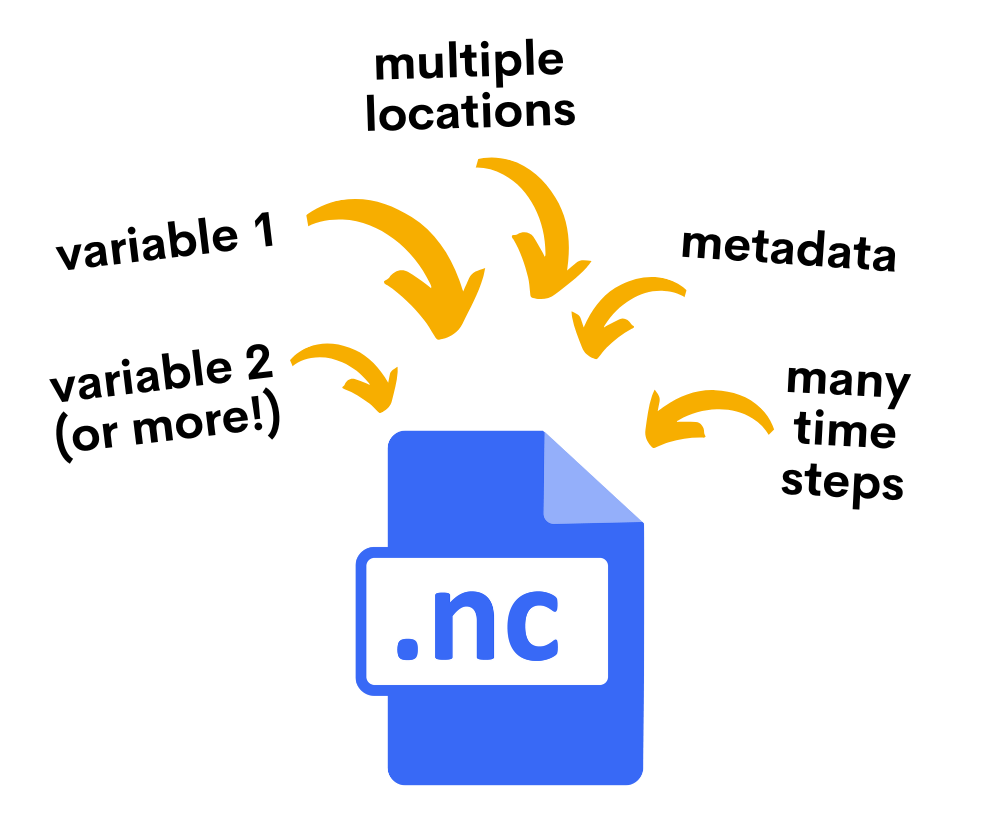
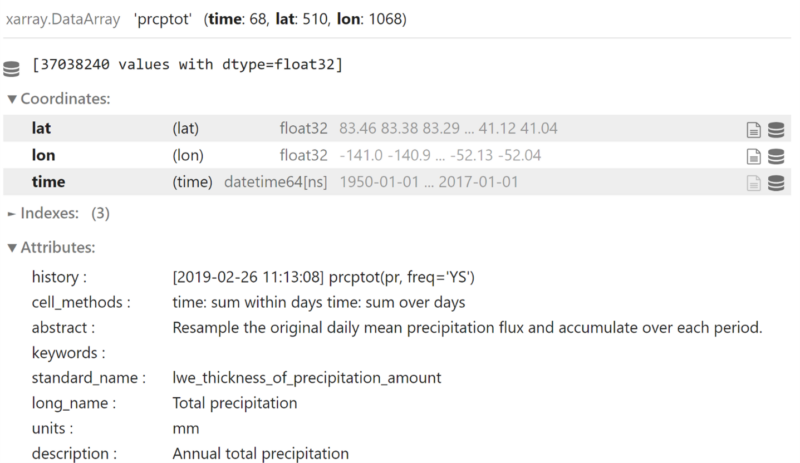
Full Tutorial
After opening and viewing the contents of a netCDF file, users will very likely want to perform additional custom analyses (e.g. computing a custom metric, such as “number of days per year that are above 33°C relative across a specific region”) prior to creating custom data visualizations (i.e. maps, graphs, and tables).
Again, PAVICS contains several built-in tutorials that showcase how to program these types of analyses and visualization commands. It is outside the scope of this article to explain how to develop these types of programs; however, the following code example demonstrates how one might approach this type of custom analysis.
The code block pasted below extracts and analyzes climate data from a netCDF file for a specific region, in this case, Terra Nova National Park. It begins by loading a shapefile that contains the boundary of the park and reprojects it to a standard geographic coordinate system. The program then accesses a climate dataset (specifically maximum temperature projections from one downscaled global climate model) from a specified URL and subsets the data to match the park’s geographical boundaries. Using the xclim library, it calculates the number of days per year where the maximum temperature exceeds 33°C. The program further computes two types of averages: one that averages the data spatially across the park, and another that averages the data temporally over the period from 2051 to 2080. Finally, it visualizes the data on a geographic map using a quadmesh plot, which includes a color scale to represent the results and a basemap for geographical context.